|
Darwin
1.10(beta)
|
|
Darwin
1.10(beta)
|
This tutorial is designed to give you a basic flavour for the design and use of the common features of the Darwin libraries. It assumes you are familiar with C++ and the Linux development environment.
The tutorial is divided into a number of parts, each exploring a different library within the Darwin framework. The parts are:
Source code for all the tutorials is included in the projects/tutorial directory.
This tutorial steps you through creating your first application using the Darwin framework under Linux—the application will simply print "hello world" using Darwin's logging system (see Messages, Warnings and Errors). It assumes that you have successfully downloaded and compiled the Darwin libraries (see Downloading and Installing). Throughout the tutorial we will assume that $DARWIN is the location (i.e., directory) where Darwin is installed.
Create a file called helloworld.cpp containing the following code:
Create a file called Makefile containing the following:
-stdlib=libstdc++ to the end of the g++ command. You may also need to add -D__APPLE. On Linux systems (e.g., Ubuntu) you may need to add -D__LINUX__.The following shell commands will build the helloworld application and run it with some different settings.
Darwin provides a number of mechanisms for configuring applications from the command-ine. This is a very basic tutorial to get you familiar with the two main mechanisms—commandline options and XML configuration. Be sure to read the commandline processing documentation (Command Line Processing) and the configuration module documentation (Configuration Manager).
The following code is an example of how to configure a Darwin application either via the commandline options or an XML configuration file.
Note that there is no semicolon (";") after the intermediate commandline processing macros.
Create the following configuration file called "myConfig.xml":
Compile the code and then run with the following options:
This tutorial demonstrates how to make use of Darwin's XML utilities for serializing objects to disk. Any class derived from drwnWriteable will have access to read and write methods that read and write XML files to disk. The derived class will need to override the load and save methods.
We will begin by creating a serializeable class that holds a person's name and age.
Of course, to be useful this class would need to implement a number of other methods (for example, member variable accessors). However, this lean implementation is enough for our purposes in this tutorial.
We now create a class that aggregates many people into a single group.
Finally, we create an application that populates a group of people and writes the group to disk.
Compile this program (you will need to write the appropriate makefile), run it, and examine the output.
Often we wish to store large volumes of binary data between sessions where XML files may be too inefficient or memory intensive. The drwnPersistentStorage class provides a simple mechanism for storing and managing the reading and writing of multiple data records for such situations.
Suppose the data we wish to store is of type std::vector<double>. The first step is to provide a drwnPersistentRecord wrapper:
We can now easily read, write and modify data records.
The data is stored permenantly when the application closes and can be read later by re-opening the same storage file.
The drwnML library implements a number of basic machine learning and pattern recognition algorithms. These are explored in the following tutorials.
Darwin provides functionality for estimating the parameters of, sampling from, computing sample likelihoods, and conditioning on multi-dimensional Gaussian and mixture of Gaussian probability distributions. The following example shows how to define a two-dimensional Gaussian and then condition on the first variable to produce some one-dimensional Gaussian distributions.
When performing multiple repeated conditioning on large Gaussians, creating an intermediate drwnConditionalGaussian object is more efficient since it will cache some intermediate calculations.
Exercise. Extend the above example to produce samples from the conditional distribution.
The drwnFeatureTransform class defines the interface for supervised and unsupervised feature vector transformations, e.g., Fisher's LDA (supervised) or PCA (unsupervised). For many machine learning algorithms pre-processing features to have zero mean and unit variance will result in better numerical stability of the algorithm. This tutorial shows such an example.
Once the feature transformation has been learned it can be applied to arbitrary feature vectors (i.e., not just those that where used during learning). For example,
The same data can be trasnformed via Principal Component Analysis (PCA). In the following example we set the number of output dimensions to 5.
We can also perform PCA on a transformed feature space, e.g.,
Here the feature vectors are automatically transformed via the drwnSquareFeatureMap before learning or applying PCA.
Classification and regression and the two most fundamental tasks in supervised machine learning. Both tasks take as input a feature vector and produce as output a prediction. In the case of classification the prediction is a label from a small discrete set, whereas in the case of regression the prediction is a real-valued number.
The following example shows how to use the drwnTLinearRegressor class for extrapolating a straight-line through some data points.
The following code shows a how to use the drwnClassifier class.
The feature mapping used by the multi-class logistic classifier can be easily changed. For example, to augment the square of each feature use
The code snippet above shows analysis of the results on the training set by printing a confusion matrix. Analysis should really be done on a separate hold-out set. In addition to the confusion matrix you may be interested in plotting a precision-recall curve (see drwnClassificationResults).
The drwnPGM library provides infrastructure for representing and manipulating graphical models such as Bayesian Networks and Markov random fields. This section gives you an overview of some of the functionality of the library. We begin by demonstrating construction of a factor over a set of discrete variables and then move onto a simple energy minimization example followed by a Viterbi decoding example. The tutorial ends with some advanced topics, specifically, creating factor operations and sharing factor storage.
A factor is a table containing a value for each assignment to a set of random variables. The following code shows how to construct a factor psi over two variables A and B.
When developing algorithms we often wish to perform a core set of operations on the factors in the graphical model. These operations are encapsulated by the drwnFactorOperation class and classes derived from it. The following code snippets show some simple operation use-cases. More complex examples are demonstrated in subsequent sections of this tutorial.
Consider the following energy minimization problem over discrete random variables x,
![\[ \textrm{minimize} \; \sum_{c} \psi_c(\mathbf{x}_c) \]](form_9.png)
A large number of algorithms have been proposed for (approximately or exactly) solving this problem depending on the structure and properties of the energy function. Darwin implements a number of these algorithms and in this tutorial we demonstrate using the ICM algorithm (Besag, 1975).
We first construct a factor graph representation of the energy function.
Now let's try use ICM to compute the minimum-energy assignment:
For low treewidth graphs we can use the junction tree algorithm, which first triangulates the graph, to compute the exact MAP assignment:
We now provide an example of implementing the Viterbi algorithm using the factors and factor operations available in the library. The Viterbi algorithm is a method for finding the most likely sequence of states in a Markov model. For this tutorial we will assume that our model is a chain defined over T time steps by
![\[ P(\mathbf{x}) \propto \prod_{t = 1}^{T - 1} \Psi_{t}(x_t, x_{t+1}) \]](form_10.png)
The following code implements the viterbi algorithm on a sequence of factors. Note that this implementation is far from optimal and intended for illustrative purposes only. Practical implementations of the algorithm would store the argmax during the forward pass to avoid the reduction computation in the backward pass. The normalization of the messages is required to avoid overflow. Some viterbi implementations avoid this by performing the computation in log-space. Try this as an exercise.
The following main function provides an example of setting up a Markov chain and calling the viterbi code.
In this tutorial we demonstrate how to write a new factor operation that computes the dot-product between two factors, i.e., given 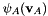 and
and 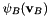 with
with 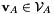 and
and 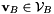 will compute
will compute
![\[ \sum_{\mathbf{v} \in {\cal V}_A \cup {\cal V}_{B}} \psi_A([\mathbf{v}]_A) \psi_B([\mathbf{v}]_B) \]](form_15.png)
We first define the operation class. Most factor operations produce another factors and so should be derived from the drwnTableFactorOp base class. However, our factor computes a real number which we will return through a results pointer. The class is defined below. It contains a pointer to each of the input factors and the mapping between corresponding entries in those factors.
The constructor computes the union of the variables in each factor and the mapping between factors and this variable superset. Since the mappings are initialized on construction repeated execution of the operation (say, in an iterative algorithm where the contents of the factors are changing) are relatively cheap.
The execute method performs the dot-product operation. In our case its behaviour is to virtually expand each factor to be over the full label space of the union of variables by replicating entries. It then sums the product of corresponding entries in these two virtual factors.
The following main() function shows example invocations of the the DotProduct class. The tutorial project contains a more elaborate example.
Sometimes the same values appear in many different factors (i.e., over different variables) or factors are used to store temporary results at different times in the execution of an algorithm. In these situations the drwnTableFactorStorage class can be used to reduce memory footprint and limit repeated memory allocations and deallocation. The following code snippet provides a simple example of sharing values in many factors.
You are now ready to start your own project using Darwin or explore some of the existing applications and projects. If you are interested in contributing to the library email stephen.gould@anu.edu.au.
 1.8.6
1.8.6