S. Hamid Rezatofighi
PhD Student of Engineering & Computer Science


Home |
Research |
CV |
News |
Contact |

ANU |
RSISE |
CSIRO |
Login |
Recent projects

SEGMATION OF VASCULAR STRUCTURES
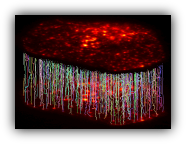
MOTION ANALYSIS OF HAIR FOLLICLE STEM CELLS IN VIVO LIVE IMAGING

Stem cells and the microenvironment in which they reside - the so called niche - are central for the development and regeneration of all our organs, and their deregulation leads to a disease state. Despite the key relevance of stem cell niches and their conserved features, the dynamic interactions between stem cells and the niche are still not well understood. The aim of this project is to assess these dynamic interactions in order to understand how stem cells and the niche contribute to tissue regeneration and what goes awry during disease states such as cancer using the murine skin hair follicle as a model system.
The project is in close collaboration with Yale School of Medicine, USA, Helmholtz Association of German Research Centers and the Technical University of Munich, Germany.
Despite significant technical advances made in automatically segmentation of tubular structures, the vessel segmentation still remains a challenging task in many practical applications due to the complex nature of the vascular networks. These networks usually includes numerous branching and bifurcations in where many segmentation algorithms fails to work properly. Moreover, the vessels may not be distinguished well from its surroundings due to low image contrast, noise, vicinity to other structures with similar brightness and other pathological reason.
This project provides a novel perspective to the vascular centerline tracking and segmentation problem using Bayesian approach. The project is in close collaboration with Computer Aided Medical Procedures & Augmented Reality Group in the Technical University of Munich, Munich Germany.
Quantitative analysis of the dynamics of tiny cellular and subcellular structures in time-lapse cell microscopy sequences requires the development of a reliable multi-target tracking method capable of tracking numerous similar targets in the presence of high levels of noise, high target density, maneuvering motion patterns and intricate interactions.
In this project, we focus on tracking these structures in different biological applications based on Bayesian frameworks. This project is in close collaboration with Garvan Institute of Medical Research, Australia

MULTI-TARGET TRACKING IN TIME-LAPSE CELL MICROSCOPY SEQUENCES
Past projects

REALISTIC SIMULATION OF FLUORESCENCE MICROSCOPY SEQUENCES
Since generation of reliable ground truth annotation of fluorescence microscopy sequences is usually a laborious and expensive task, many proposed detection and tracking methods have been evaluated using synthetic data with known ground truth. However, differences between real and synthetic images may lead to inaccurate judgment about the performance of an algorithm.
In this project, we present a framework for generating realistic synthetic sequences of fluorescence microscope through simulation of the image formation process and accurate measurement and dynamic models. We believe that the sequences generated using this framework appropriately reflect the complexities existing in real sequences. Therefore, the sequences can be an apt evaluator for detection and tracking methods.

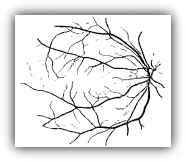
Inspection of optic fundus photographs may be conducive to diagnose and monitor the progress of general diseases such as diabetes, hypertension, arteriosclerosis, cardiovascular diseases, stroke and eye diseases like retinopathy of prematurity. Thus, the measurement and analysis of retinal blood vessels is of diagnostic value for a wide range of pathological states. However, manual analysis of the complex retinal blood vessel trees in fundus images is a tiresome and laborious task, and as the number of images increases, it may even be impossible.
Segmentation of the vessels from the background is an initial requirement for the automatic assessment of retinal images. In this project, we present novel supervised and unsupervised methods for the automated segmentation of blood vessels.
RETINAL VESSEL SEGMENTATION IN FUNDUS IMAGES
AUTOMATIC RECOGNITION OF WHITE BLOOD CELLS IN HEMATOLOGICAL IMAGES
Recognition and inspection of white blood cells in peripheral blood can assist hematologists in diagnosing diseases like AIDS, leukemia, and blood cancer, making it one of the most salient steps in hematological procedures. This analysis can be accomplished by automatic and manual approaches. However, manual scrutiny of blood cells by experts is tedious and susceptible to error. Therefore, an automatic system based on image processing techniques can help the hematologists and expedite the process.
In this project, our main purpose is to develop a new algorithm based on image processing methods to detect and classify different types of white blood cells in the peripheral blood.

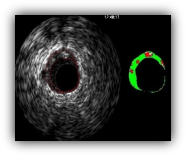
Grayscale intravascular ultrasound (IVUS) provides real-time and high-resolution tomography visualization of the coronary arteries. Lumen and vessel dimensions and the distribution of different plaques can be analyzed through inspecting images driven by IVUS imaging modality.
Applying the informative rich IVUS frames of each patient and performing manual analysis of cross section area images is not only time-consuming but also prone to the inter-observer and intra-observer variability. Furthermore, the clear identification of nonclassified, i.e. fibrous or rich lipid, plaques are especially difficult. Thus, developing a device capable of characterizing plaque components by IVUS images automatically is of great importance. In this project, a new tool capable of automatic IVUS image analysis is proposed.
AUTOMATIC ANALYSIS OF IVUS IMAGES: FROM BODER DETECTION TO PLAQUE CHARACTERIZATION

DETECTION AND SEGMENTATION OF HEPATIC HEMANGIOMA IN ULTRASOUND IMAGES
Hemangioma is an unusual formation of blood vessels in the skin or internal organs which is considered a benign (nonmalignant) congenital complication. Some hemangioma lumps appear and form in the liver causing problems for the affected individual. This lesion is the most common type of hepatic tumors. Wrong detection of such a lesion can cause an unnecessary surgery for the patient whereas an on time and exact detection of this disease can enable the treatment of it with low risk and less cost. The chances for early clinical assessment of hepatic hemangioma by employing biopsy is often small and almost impossible because of its hyper vascular structure. Ultrasound imaging and scanning of the liver are, therefore, commonly utilized as the routine methods of liver tumor detection and diagnosis.
In this project, an automatic method for detection and segmentation of hepatic hemangiomas in ultrasound images of the liver is proposed.
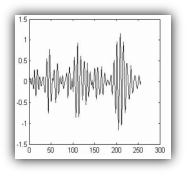
BIOMETRICS USING VISUAL EVOKED POTENTIALS
Biometrics is the technique of uniquely recognizing a person among a group of people. It is usually performed based on one or more of humanís intrinsic physical or behavioral traits such as fingerprints, iris, speech, face, etc. These traits, however, can be altered or manipulated either intentionally or accidentally. For this reason, researchers have been investigating for new biometrics in the last couple of years which can satisfy the important conditions of (a) being unique for any individual; (b) not changed or be forged easily; and (c) being not so complex for analyzing. Considering these conditions, such biometrics as electrocardiogram (ECG) and electroencephalogram (EEG) has been proposed for person identification.
In this project, the feasibility of Visual Evoked Potential (VEP) in the gamma band of EEG signal, as a physiological trait, is studied, and used to identify individuals in a group of people.